|
Manduca sexta Larvae Exhibit Rhythms in Feeding Behavior and Rhythms in Gene Expression of the Circadian Gene period Donovan Waters, Howard Community College |
Abstract
The tobacco hornworm (Manduca sexta) is considered an agricultural pest due to its predation on agriculturally significant Solanaceous plants, such as tomatoes and tobacco, making the behavioral and molecular mechanisms underlying its feeding behavior of great interest to agriculture. Circadian rhythms are daily biological rhythms endogenous to an organism that can be reset by environmental cues and have been shown to influence the behavioral output of organisms. The molecular basis for these approximately 24-hour rhythms is a transcriptional/translational feedback loop in individual cells, involving a gene called period (per). While circadian rhythms have been shown to be functional in the adult Manduca sexta and to affect behavior, the presence of a functional circadian clock has not yet been described in the larvae. In this study, we explored whether the tobacco hornworm exhibits rhythms in feeding behavior and whether the circadian gene per is rhythmically expressed. When we tested larvae at different times of day, we found that there were significant differences in the amount of leaf they consumed at different timepoints. In addition, when tissue samples were harvested at these same time points, the expression level of per also significantly differs at different times of the day. Our data is indicative of the tobacco hornworm exhibiting rhythmic feeding behavior and circadian gene expression, but further investigation will be necessary to determine if their feeding behavior is indeed modulated by circadian rhythms.
Introduction
Mammals, fish, insects, various plants, and even some cyanobacteria [1]-[5] exhibit oscillations in their biological systems and behaviors over the period of a terrestrial day and night. The word circadian itself is Latin for about (circa) a day (dies) [6]. Genetic and molecular signal underpinnings for this cycling have been explored from many angles in numerous model organisms. There are genetic transcriptional and translational feedback loop (TTFL) mechanisms that account for much of this cyclicity [7], which includes various genes such as period (per), timeless (tim), and cryptochrome (cry) [8], [9]. The molecular clock then affects metabolic physiology downstream [10]-[12], such as insulin production. The desynchronization of circadian rhythms has been shown to cause impairment of metabolic processes in several adult insects, including the standard model insect Drosophila melanogaster [13].
The rhythmic nature of an organisms’ internally driven (endogenous) biological timing can be synchronized by external (exogenous) stimuli. This process is called entrainment. The most well understood of these entrainment influences are light, temperature, and metabolism of food, collectively referred to as zeitgebers, the German word for “time givers” [8], [9], [14]. It has been shown that observable feeding behavior will affect and be affected by an organism’s daily biochemical oscillations [14] and similarly, conservation of circadian mechanisms over evolutionary time would synchronize between a pollinator or consumer and the plants upon which it feeds [15].
The tobacco hornworm (Manduca sexta) in its larval stage is known to be an agricultural pest and feeds primarily on Solanaceous plants such as tomato and potato [16]. Its oligophagy, specific food preference, for Solanaceous plants has been well described [17]. After M. sexta hatches from its egg, which is laid on a Solanaceous leaf, it rapidly develops through five instar larval stages in approximately two weeks. In the 5th instar, M. sexta are entering their final larval growth phase and must eat voraciously to reach their threshold body weight before pupation [18], but the molecular drivers of their feeding behavior have not been well characterized. Other endogenous and exogenous factors that are synchronous with or after feeding behaviors remain to be discovered in detail, and effective management of a pest species requires an understanding of its physiology.
Robust circadian rhythms have been shown to control rhythmic physiology in the adult M. sexta, the hawkmoth. Hawkmoth express the per gene in their antennae, which are used to sense the presence of dietary phytochemicals (bioactive plant chemical) [19]. In addition, responsiveness to floral scent is modulated by the circadian clock in adult M. sexta [15], but the presence of a function circadian clock has not yet been described in the larvae. In previous research from our lab, per gene expression has been found in the midgut of 5th instar M. sexta [20], but it is not yet known if this expression is rhythmic. It has also been unclear whether the feeding behavior of 5th instar larvae is continuous or rhythmic, and if its feeding is rhythmic, is it then related to the whether its rhythmic feeding is controlled by circadian oscillation.
Our study seeks to investigate whether M. sexta larvae exhibit rhythmicity in feeding behavior and whether they have a functional circadian clock. Therefore, our team performed an experiment to observe feeding behavior on tomato leaves to observe whether leaf consumption varied by time of day. We also assessed the per expression levels by quantitative polymerase chain reaction (qPCR) at the same behavioral timepoints to see if per levels fluctuated at different times of the day.
Methods
Caterpillar husbandry
Tomato plants were grown in the HCC greenhouse for caterpillar feeding. After the Spring 2021 cohort, tomato plants were given fertilizer to improve nutritional value and tomato plant usage was rotated after each harvesting to prevent plant stress reactions from affecting nutritional value of leaves.
Manduca sexta eggs were sourced from Carolina Biological Supplies. Rearing began with placing clusters of eggs on petri dishes with lids in a controlled environment at 22-24ºC. Daily checks for larvae health and instar (periods between molts) progress were conducted. Instar progress was determined by a combination of physical features and the presence of head capsule shedding. New leaves were provided daily following the removal of fecal matter and old food. Rearing methods continued until larvae reached 5th instar pre-molt. The Fall 2021 cohort was reared entirely in a 16:8 LD (light/dark) cycle, while the Spring 2021 cohort was placed in a 16:8 LD when molted to 4th instar.
Behavioral feeding paradigm
Cohorts were tested for feeding behavior at different zeitgeber times (ZT): ZT 1 & 13 (Spring 2021) and ZT 3 & 15 (Fall 2021). ZT 0 is the beginning of the light period of the cycle. Time points were chosen 12 hours apart with the first time point being close to the start of the light portion of the cycle (ZT 1 & 3) and the second being close to the end of the light portion of the cycle (ZT 13 & 15). A two-hour shift between Spring and Fall time points allowed for better definition of the rhythmicity of per expression levels. Shortly before cohort ZT designations, the larvae were removed from petri dishes to be weighed as well as new leaves provided for feeding. Larvae were left to feed for one hour. After one hour, the larvae and leaf were each weighed again, and mass changes were calculated.
Tissue harvesting
Following the end of behavioral testing, larvae were placed in ice baths for approximately 15 minutes for anesthetization. Dissections began after anesthetization was complete. The dissection kits used were sterilized with 95% ethanol and cleaned with RNase-free wipes to prevent RNase or nucleic acid contamination of tissue samples harvested from the larvae. Incisions were made starting behind the first pair of legs down to the midsection for removal of a portion of the midgut. Midgut samples were suspended in RNAlater (Thermofisher) to maintain stability of mRNA. Samples were kept in RNAlater at 4ºC overnight, after which they were stored at -80ºC.
RNA extraction and cDNA synthesis
Molecular analysis of samples began with the extraction and purification of mRNA from midgut samples as outlined in RNeasy Mini Kit [1] manual (Protocol: Purification of Total RNA from Animal Cells Using Spin Technology, page 27). Deviation from protocol occurred during the centrifugation of collection tubes by removing genomic DNA (gDNA) with an eliminator column. Additionally, all steps in the protocol that called for 15 seconds of centrifugation was increased to 30 seconds due to instrument limitations and 39.2 mM dithiothreitol (DTT) was added to the RLT buffer to improve lysing. Sample RNA purity was verified using a Nanodrop with A260/280 and A260/230 ratios. The 230, 260, and 280 represent the wavelengths of light (nm) at which absorbance was measured. To ensure that the nucleic acid extracted was RNA and not DNA a A260/280 ratio of ~2.0 is required [2]. To ensure that there isn’t protein or other reagent contamination a A260/230 ratio in the range of 2.0-2.2 is recommended [2]. These parameters help ensure that the product isolated is pure RNA and is free of protein and other contaminants.
The extracted mRNA was converted into single-stranded complementary DNA (cDNA). The cDNA synthesis protocol was used as outlined in the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) [3]. From the kit, the provided Gapdh RNA was used as a positive control for efficacy of the cDNA synthesis and the oligo (dT)18 primers were used for the synthesis reaction. The cDNA synthesized in this protocol was used as a template for quantitative polymerase chain reaction (see next section).
Quantitative polymerase chain reaction (qPCR)
Our qPCR was conducted on cDNA samples from the Spring caterpillar cohort as outlined in [13] using the SsoAdvanced Universal SYBR Green Supermix kit (Bio-Rad). qPCR was used for the relative measurement of gene expression levels of actin and per. The actin cDNA was used as the reference gene, as it is constitutively expressed in the tissues examined [4]. The gapdh cDNA is used as a positive control in qPCR as well due to its early amplification as an indicator of successful replication of cDNA in the previous step. Primers for gapdh were provided by the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). Primers for actin and per were designed and tested by previous URSC research students [13], [21]. Reactions containing no cDNA template, no template control (NTC), were performed for quality control. All per and actin reactions were performed in triplicate. Product specificity and product size was confirmed with both sets of primers.
Cycle threshold (CT) is the number of thermal cycles of amplification that it takes for the fluorescence signal of SYBR green bound to cDNA to cross the threshold. As cDNA is amplified, the fluorescence signal increases until all of the nucleotides are utilized in the test tube. The threshold is set at an arbitrary value within the linear amplification of fluorescence signal to allow for consistent comparison between samples. In order to examine gene expression changes in per, we compared its amplification to actin’s amplification by calculating the difference in CT value between them, delta (𝝙) CT. To find 𝝙CT for a sample, we subtracted the actin CT from the per CT. A higher 𝝙CT signifies lower gene expression, whereas a lower 𝝙CT signifies higher gene expression.
Statistical analysis
Results from Spring 2021 and Fall 2021 cohorts’ leaf consumption data were analyzed using R software (R Project Core Team). Testing for normality was completed first by using a Shapiro-Wilk normality test. Homogeneity of variance was then examined by using both Bartlett’s test and Fligner-Killeen’s test. The Spring 2021 cohort’s qPCR results were analyzed through the comparison of 𝝙CT values between time points ZT 1 & 13 for significant difference in expression levels using Welch Two Sample T-test. Significant differences in leaf consumption for both cohorts were also determined using the Welch Two Sample T-test. A p-value of less than 0.05 was considered significant.
Results
Behavioral feeding test results
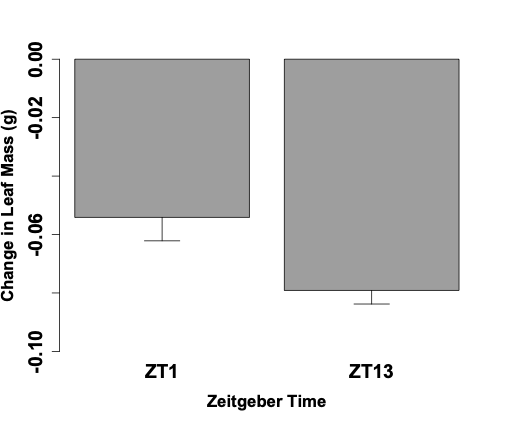
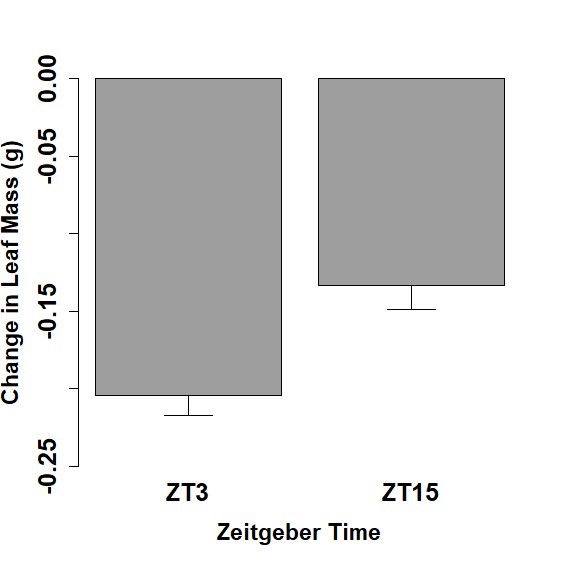
Feeding behavior was examined for a 1-hour period and the change of leaf mass over that period was calculated for both the Spring and Fall cohorts of 5th instar caterpillars. Mean leaf mass changes and standard error of the mean for the Spring cohorts were measured at -0.054 ± 0.021 g for ZT 1 and -0.079 ± 0.012 g for ZT 13 (Figure 1). Fall cohort mean leaf mass changes and standard error of the mean were measured at -0.205 g ± 0.012 for ZT 3 and -0.133 ± 0.015 g for ZT 15 (Figure 2). Caterpillars tested at ZT 1 exhibited significantly less leaf consumption than caterpillars at ZT13 (p <0.05). Caterpillars tested at ZT 15 exhibited significantly less leaf consumption than caterpillars at ZT 3 (p <0.05). Furthermore, when all the time points tested are considered, our data show that the caterpillars consumed more of the leaf in the time points closer to the middle of the light period (ZT 3 and ZT13) in relation to their counterpart time point closer to the light/dark transition (ZT 1 and ZT 15).
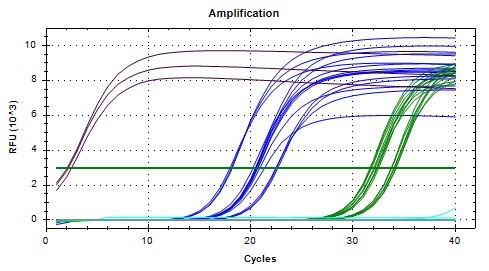
Analysis of qPCR results
In order to investigate per expression at different timepoints, qPCR analyses were conducted for midgut samples of the Spring cohort. Due to the number of samples and replicates needed, two separate qPCR analyses were performed. All the samples for a given time point (ZT 1 or ZT 13) were analyzed at the same time. The SYBR green fluorescence amplification graphs for both are shown in Figures 3 and 4. Early amplification of gapdh cDNA was seen as expected due to high concentration of starting mRNA, followed by actin (reference gene), and then per (Figures 3 & 4). The NTC replicates for both analyses did not have appreciable amplification as expected. The melting curves (not shown) further verified that only a single qPCR product was produced for each gene analyzed at the expected melting temperature. A threshold relative fluorescence units (RFU) level of 3 x 10^3 was set on both graphs for consistency as it was in the linear range of amplification for all samples analyzed.
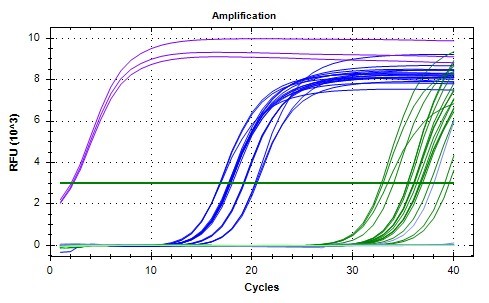
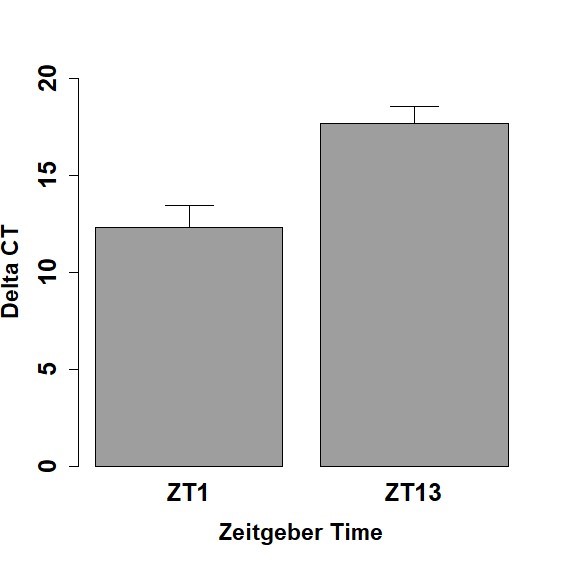
For each 5th instar caterpillar midgut sample, the mean CT value for per and mean CT value for actin were calculated from the three replicates. The overall mean 𝝙CT and standard error of the mean for all the caterpillars tested at each time point (ZT 1 and ZT 13) were then calculated. The mean 𝝙CT for Spring midgut samples was 17.70 ± 0.86 for ZT 13 and 12.34 ± 1.09 for ZT 1, respectively (Figure 5). Lower 𝝙CT values indicate higher gene expression of per in tissues. The expression level of per at ZT 1 was significantly higher than the expression level of per at ZT 13 (p <0.05). This demonstrates that per expression levels differ at different times of day, indicative of rhythmicity.
Discussion
Through the experiments conducted in this paper, we sought to determine the presence of rhythmic feeding behavior and gene expression in the 5th instar Manduca sexta larvae. To find evidence of a functioning circadian clock, per expression and feeding behavior were quantified over different time points throughout the light period. Previous research in the adult hawkmoth has shown evidence that circadian rhythms influence pollination behavior [15]. As Manduca sexta larvae are known as pests for Solanaceous plants, establishing whether circadian rhythms are functional and influencing behavior in the larval stages of this organism may provide assistance in the management of it.
The larval stages of Lepidopteran species seek to consume maximal amounts of food in preparation of pupating. Our analysis of feeding behavior at time points ZT 1, 3, 13, and 15 show significantly more feeding occurring during the time points closer to the middle part of the day for each cohort compared to the counterpart time points closer to the light/dark transition. This is surprising since the adult hawkmoth is known for being nocturnal with feeding, flight, and pollination behavior reduced in light conditions [15]. While our experiment does not preclude nighttime feeding activity, the larval feeding behavior quantified in this experiment shows significant day time feeding in the larvae. In addition, our data is the first to suggest that the larval feeding behavior or Manduca sexta is rhythmic, although rhythmic feeding behavior has been described in the midgut of another caterpillar species: the cotton leafworm caterpillar, Spodoptera littoralis [22].
Another focus of our study was to determine whether the larvae of Manduca sexta have a functional circadian clock exhibit rhythmic circadian gene expression like the adult. The midgut samples from ZT 1 and ZT 13 cohorts show significant changes in per expression at different hours of the day, but to establish a profile of rhythmicity more time points would need to be examined. In addition, these data are consistent with circadian rhythms being present in the larvae. However, persistence of these rhythms in constant darkness would need to be established before calling the rhythms circadian.
Results for feeding and per gene expression both point towards a functioning circadian clock within the 5th instar Manduca sexta larvae. However, there were limitations in the scope of the data collected. The molecular evidence for circadian rhythms in this species may be furthered examined through the processing of the ZT 3 and 15 gut samples in order to increase our sample size and expand our time points. Due to the time constraints of the project those samples have yet to be analyzed. The relative expression of per to actin during those hours may help to establish a better image of trends in expression and possible correlations between per expression and feeding behavior. Furthermore, behavioral testing at additional time points is needed to verify the robustness of our findings of rhythmicity in feeding. To expand on the results of this research, it should be further investigated as to whether the behavioral phenomenon we are seeing is a direct effect of light or indeed modulated by circadian rhythms. It is possible that the presence or absence of light may directly inhibit consumption of leaf matter. Duplication of the experiment in constant darkness may be used to determine if rhythmic feeding is controlled by the circadian clock or a direct effect of light.
Acknowledgments
In writing this paper it is important to first acknowledge the mentors through whom it was possible to complete. This project would not be possible without the guidance of Dr. Ellena McCarthy and Dr. Hannah Pie. The knowledge shared by them was invaluable. Thank you to Keighley Hayes, Zawn Nyaui, and Susan White for their help in initiating the writing of this paper as lab partners. An additional thank you is owed to Tylen Darling and Caitlyn Beckjord for providing healthy plants with which the caterpillars received nourishment.
Contact: donovan.waters@howardcc.edu, emccarthy@howardcc.edu, hpie@howardcc.edu
References
[1] P. Kim et al. Coupling the Circadian Clock to Homeostasis: The Role of Period in Timing Physiology. Endocrine Reviews, vol. 40, no. 1, pp. 66–95, Feb. 2018, doi: doi.org/10.1210/er.2018-00049.
[2] Y. Sun et al. The Molecular Evolution of Circadian Clock Genes in Spotted Gar (Lepisosteus oculatus). Genes, vol. 10, no. 8, pp. 622, Aug. 2019, doi: doi.org/10.3390/genes10080622.
[3] Y. Moriyama, T. Sakamoto, S.G. Karpova, A. Matsumoto, S. Noji, and K. Tomioka, RNA Interference of the Clock Gene period Disrupts Circadian Rhythms in the Cricket Gryllus bimaculatus. Journal of Biological Rhythms, vol. 23, no. 4, pp. 308–318, Aug. 2008, doi: doi.org/10.1177/0748730408320486.
[4] A.N. Dodd, Plant Circadian Clocks Increase Photosynthesis, Growth, Survival, and Competitive Advantage. Science, vol. 309, no. 5734, pp. 630–633, Jul. 2005, doi: doi.org/10.1126/science.1115581.
[5] Y. Xu, Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. The EMBO Journal, vol. 22, no. 9, pp. 2117–2126, May. 2003, doi: doi.org/10.1093/emboj/cdg168.
[6] “Definition of CIRCADIAN,” www.merriam-webster.com.https://www.merriam-webster.com/dictionary/circadian (accessed Apr. 17, 2021).
[7] T.S. Andreani, T.Q. Itoh, E. Yildirim, D.S. Hwangbo, and R. Allada, Genetics of Circadian Rhythms. Sleep Medicine Clinics, vol. 10, no. 4, pp. 413–421, Dec. 2015, doi: doi.org/10.1016/j.jsmc.2015.08.007.
[8] D. Sidote, J. Majercak, V. Parikh, and I. Edery, Differential Effects of Light and Heat on the Drosophila Circadian Clock Proteins PER and TIM. Molecular and Cellular Biology, vol. 18, no. 4, pp. 2004–2013, Apr. 1998, doi: doi.org/10.1128/mcb.18.4.2004
[9] E.S. Egan et al. An Extraretinally Expressed Insect Cryptochrome with Similarity to the Blue Light Photoreceptors of Mammals and Plants. The Journal of Neuroscience, vol. 19, no. 10, pp. 3665–3673, May. 1999, doi: doi.org/10.1523/JNEUROSCI.19-10-03665.
[10] M. Harrington, P. Molyneux, S. Soscia, C. Prabakar, J. McKinley-Brewer, and G. Lall, “Behavioral and neurochemical sources of variability of circadian period and phase: studies of circadian rhythms of npy−/− mice. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology, vol. 292, no. 3, pp. R1306–R1314, Mar. 2007, doi: doi.org/10.1152/ajpregu.00383.2006.
[11] N. Le Minh, Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. The EMBO Journal, vol. 20, no. 24, pp. 7128–7136, Dec. 2001, doi: doi.org/10.1093/emboj/20.24.7128.
[12] P. Crosby et al. Insulin/IGF-1 Drives PERIOD Synthesis to Entrain Circadian Rhythms with Feeding Time. Cell, vol. 177, no. 4, pp. 896-909, May. 2019, doi: doi.org/10.1016/j.cell.2019.02.017.
[13] R. Erion, A.N. King, G Wu, J.B. Hogenesch, and A. Sehgal, Neural clocks and Neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic Tissue. eLife, vol. 5, Apr. 2016, doi: doi.org/10.7554/eLife.13552.001.
[14] R.C. Northeast, V.V. Vyazovskiy, and D.A. Bechtold, Eat, sleep, repeat: The role of the circadian system in balancing sleep-wake control with metabolic need. Current Opinion in Physiology, vol. 15, pp. 183–191, June. 2020, doi: https://doi.org/10.1016/j.cophys.2020.02.003.
[15] M.P. Fenske, L.P. Nguyen, E.K. Horn, J.A. Riffell, and T. Imaizumi, Circadian clocks of both plants and pollinators influence flower seeking behavior of the pollinator hawkmoth Manduca sexta. Scientific Reports, vol. 8, no. 1, Feb. 2018, doi: doi.org/10.1038/s41598-018-21251-x.
[16] D.M. Kolodny-Hirsch, and F. P. Harrison, Yield Loss Relationships of Tobacco and Tomato Hornworms (Lepidoptera: Sphingidae) at Several Growth Stages of Maryland Tobacco. J Econ Entomol, vol. 79, no. 3, pp. 731–735, Jun. 1986, doi: doi.org/10.1093/jee/79.3.731.
[17] L.M. Schoonhoven, Loss of Hostplant Specificity by Manduca sexta After Rearing on an Artificial Diet. Entomologia Experimentalis et Applicata, vol. 10, no. 2, pp. 270–272, Jun. 1967, doi: doi.org/10.1111/j.1570-7458.1967.tb00065.x.
[18] Y. Suzuki, T. Koyama, K. Hiruma, L.M. Riddiford, and J. W. Truman, A molt timer is involved in the metamorphic molt in Manduca sexta larvae. Proceedings of the National Academy of Sciences, vol. 110, no. 31, pp. 12518–12525, Jul. 2013, doi: doi.org/10.1073/pnas.1311405110.
[19] J. Schuckel, K.K. Siwicki, and M. Stengl, Putative circadian pacemaker cells in the antenna of the hawkmoth Manduca sexta. Cell and Tissue Research, vol. 330, no. 2, pp. 271–278, Nov. 2007, doi: doi.org/10.1007/s00441-007-0471-x.
[20] M. Green, J. Jun, E. McCarthy, and H. Pie, Molecular Mechanisms for Circadian Rhythms in the Tobacco Hornworm (Manduca sexta). Journal of Research in Progress, vol. 4, pp. 24–34, 2021.
[21] A. Arhampong, M. Arminio, J. Bucksell, S. Deljookorani, B. Tegeredegn, Neuropeptide F mRNA Expression in the Tobacco Hornworm. Journal of Research in Progress, vol. 2, pp. 12-19, 2019.
[22] A. Suszczynska, M.M. Kaniewska, P. Bebas, J.M. Giebultowicz, and J. Kotwica-Rolinska, Circadian regulation of caterpillar feeding and growth, Journal of Insect Physiology, vol. 101, pp. 113–122, Aug. 2017, doi: doi.org/10.1016/j.jinsphys.2017.07.009.